In my first post about Neo4j, I explored Python modules and their dependencies. Now in this post, I am discussing how you can find the relationship between medicines, their ingredients, and their classification. But why did I choose this dataset in the first place so here is the background
Background: Currently, there is a shortage of Panadol in Pakistan, a very famous brand to cure headaches and bodily pain because the company has stopped producing it as they don’t find it viable anymore. Existing Panadol strips which used to cost Rs. 30 are even available for Rs.80 because of the lack of awareness and amazing marketing by the company. Many local websites are suggesting alternatives to Panadol. I am not a Pharma guy but I knew that in my childhood my parents used to buy Calpol and Paracetamol syrup and a few years back I watched the famous episode of Aamir Khan’s program Satyamev Jyate in which he invited a doctor who introduced the term Generic Name of the medicine. The episode went viral not only in India but Pakistan as well. To be honest it was a shocking revelation. The recent Panadol issue recalled everything and I started exploring websites to find data about medicines and their alternatives. There are many websites available but could not find one as friendly as Drugs.com. I had to equip myself with a few terms before exploring the website and finding the relevant information. Basically, I was looking for a certain brand of medicine name and the ingredients/active agent/generic name/ used in it. Once the data was available, I thought it’s a good excuse for my next post related to Neo4j.
If you want to see the demo then you can view the video below:
Development Setup
Unlike the previous post in which I used a Docker-based community edition of Neo4j, I preferred to use Neo4j for Desktop which you can download from here.
So, as I mentioned above, the goal is to find the relationship between the main ingredient a.k.a Active Agent, and the medicine itself. Bonus point, I also thought to fetch the drug’s class as well. Drugs.com’s website has all such information available. So I wrote a couple of quick scrapers to pull my required info and store it in an SQLite database. I am not gonna discuss the scraping details. I will upload it on Github and you can read the code here.
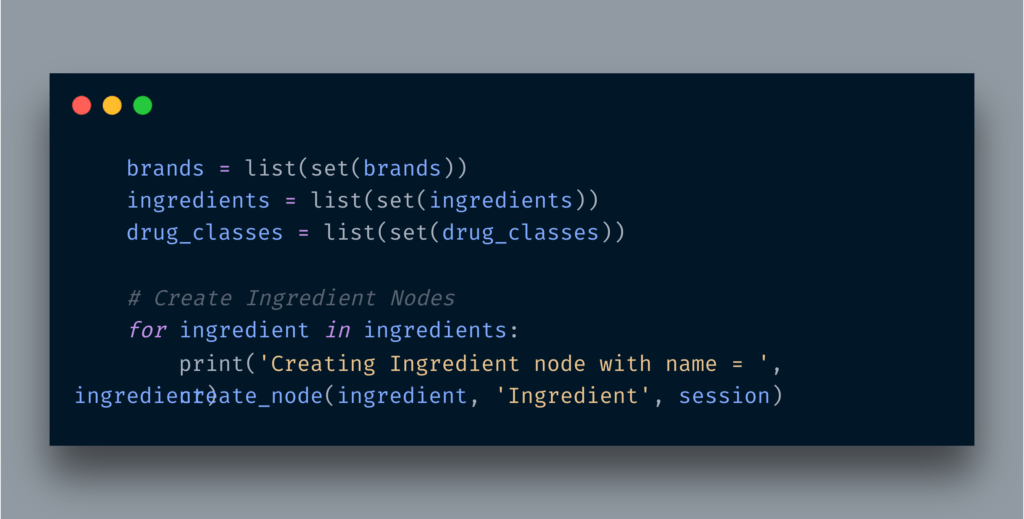
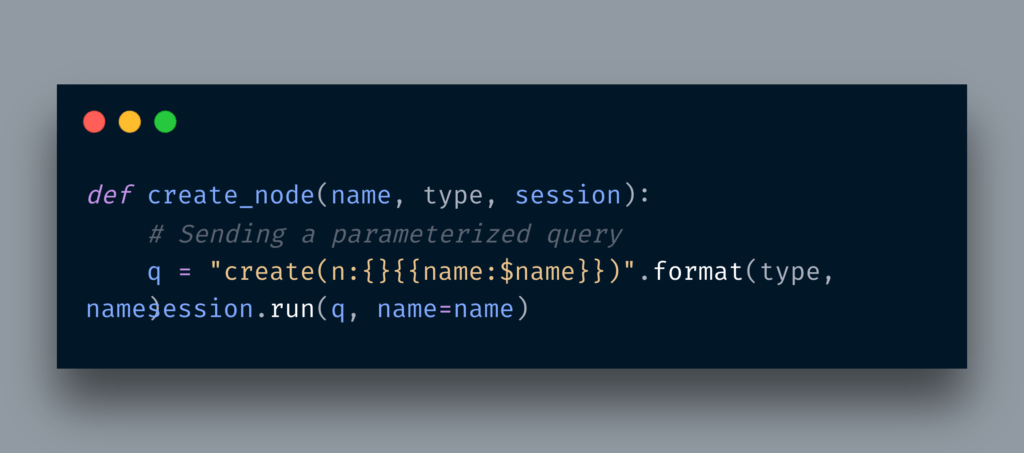
Assuming all the data available in the DB. The very first step was to pull all the Ingredient names and create their nodes.
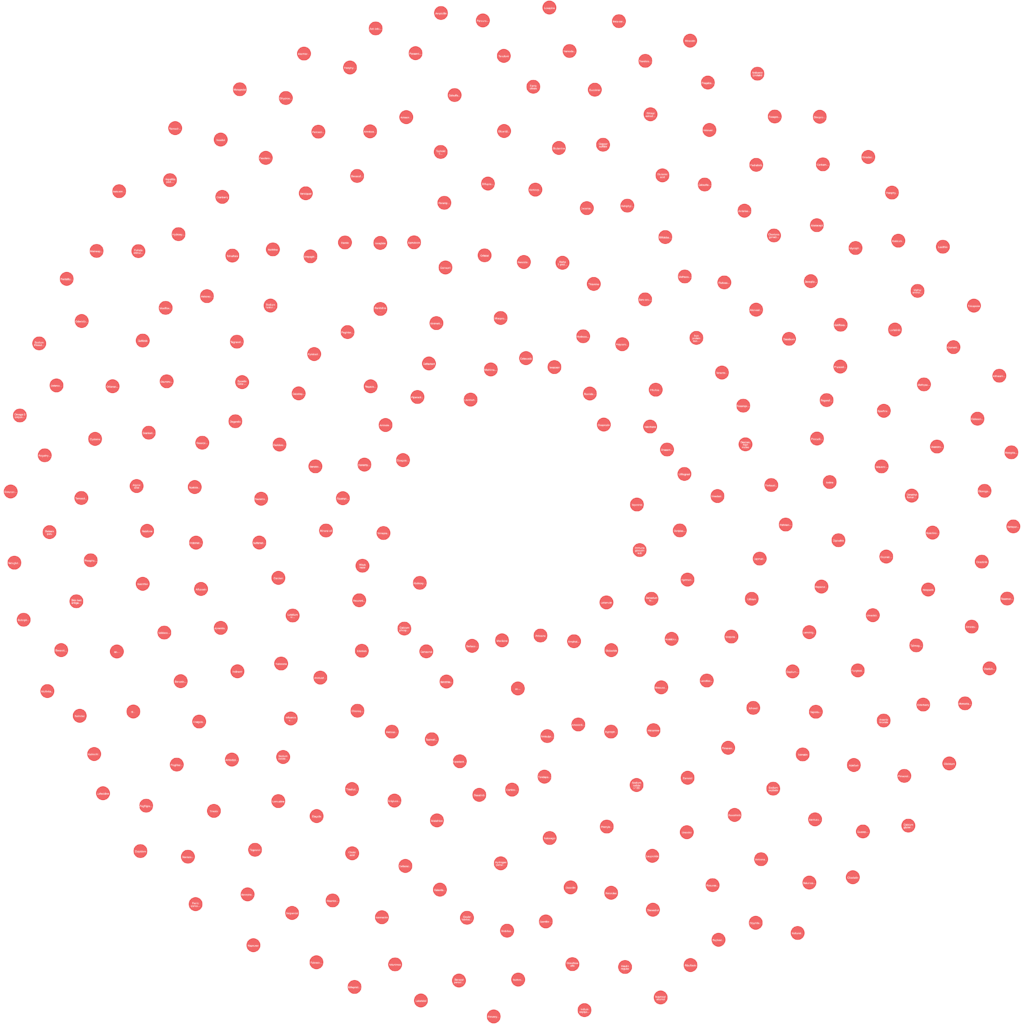
If all goes well and If I want to visualize all Ingredient nodes then they look like the below:
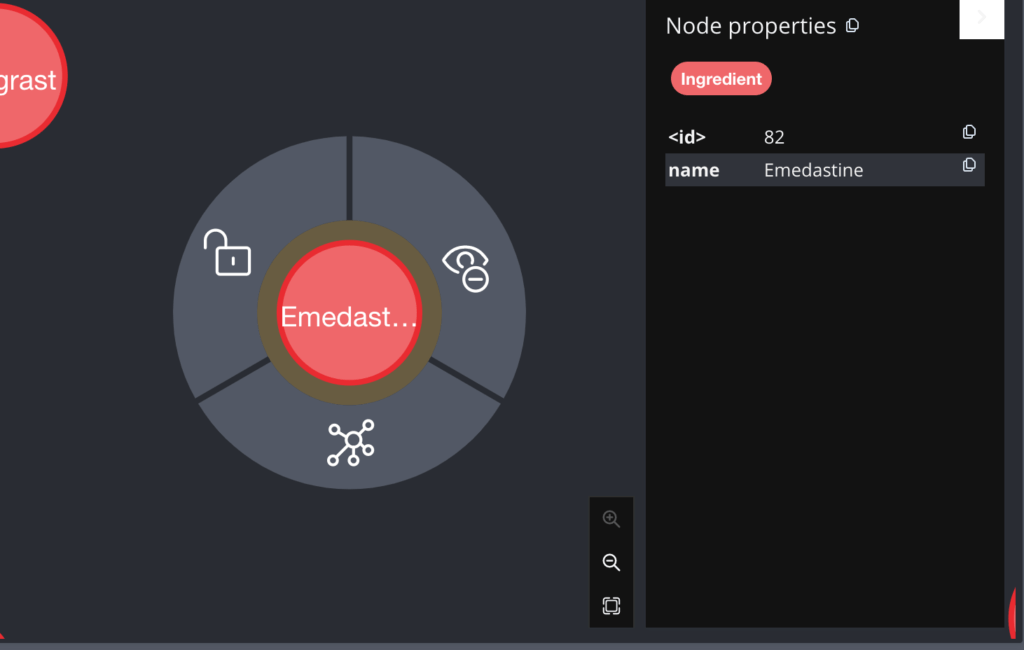
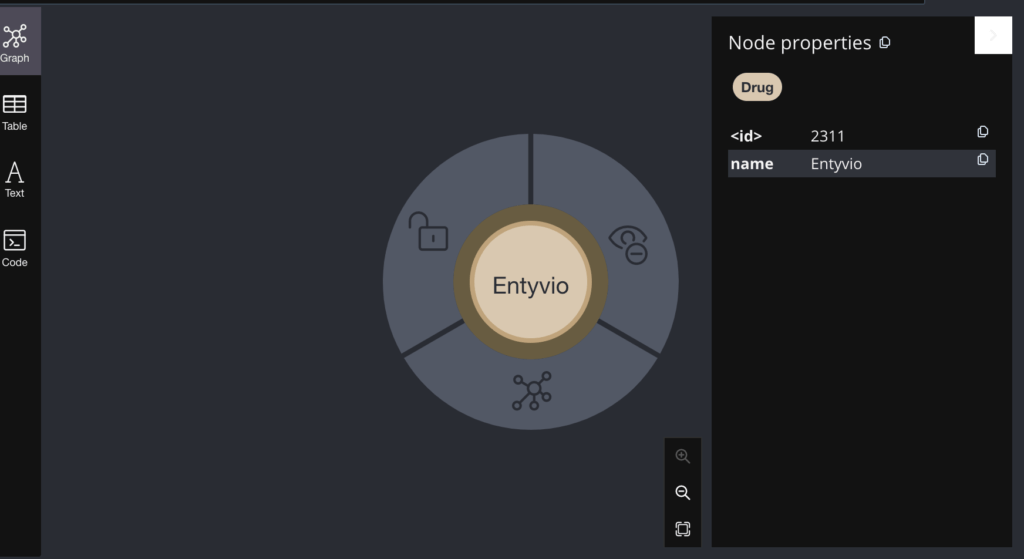
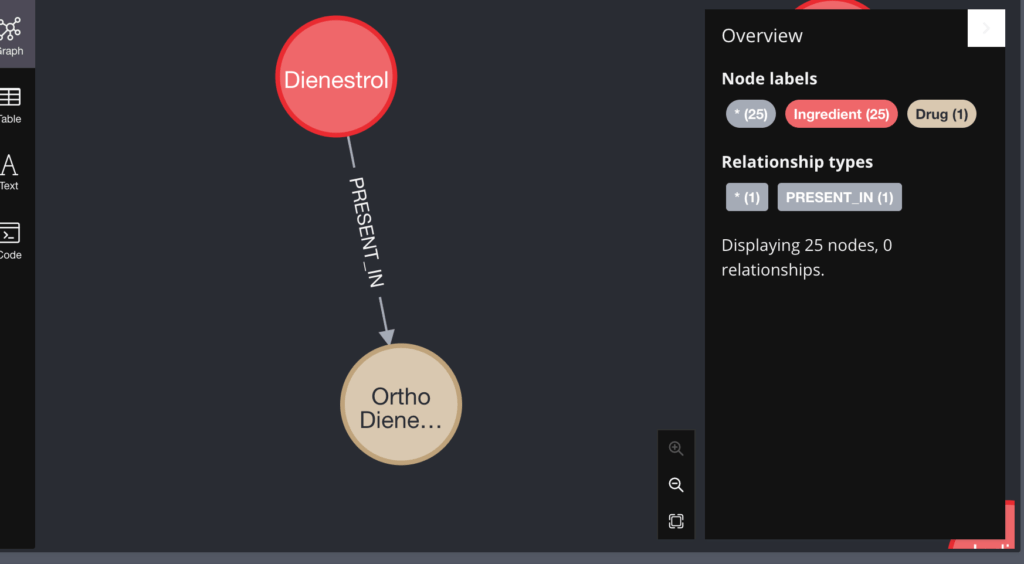
These are the initial 300 nodes only. If I want to view a single node then it looks like the below:
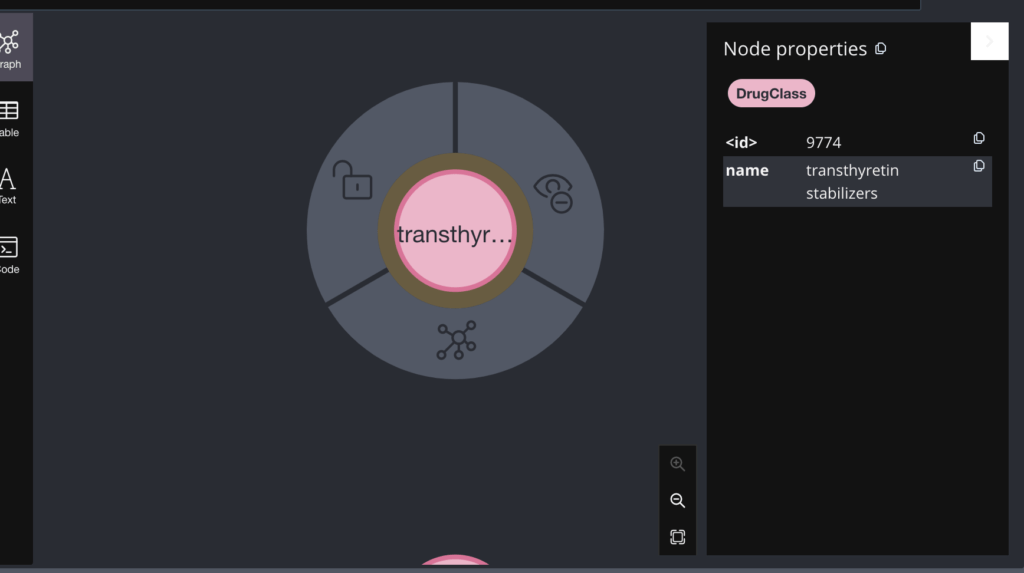
You can see both the name and ID of the node in the right pane. Similarly the Drug node and the DrugClass Node
Below is the simple routine of creating a node with a label.
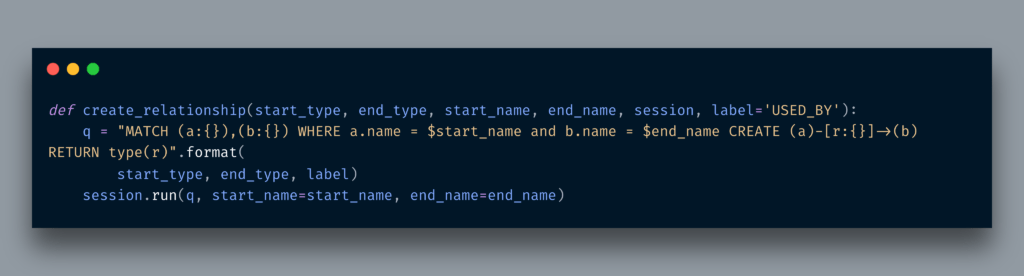
Now all nodes are available, we can now create a relationship between Ingredient and Drug first and then between Drug and DrugClass. The code for creating a relationship is given below:
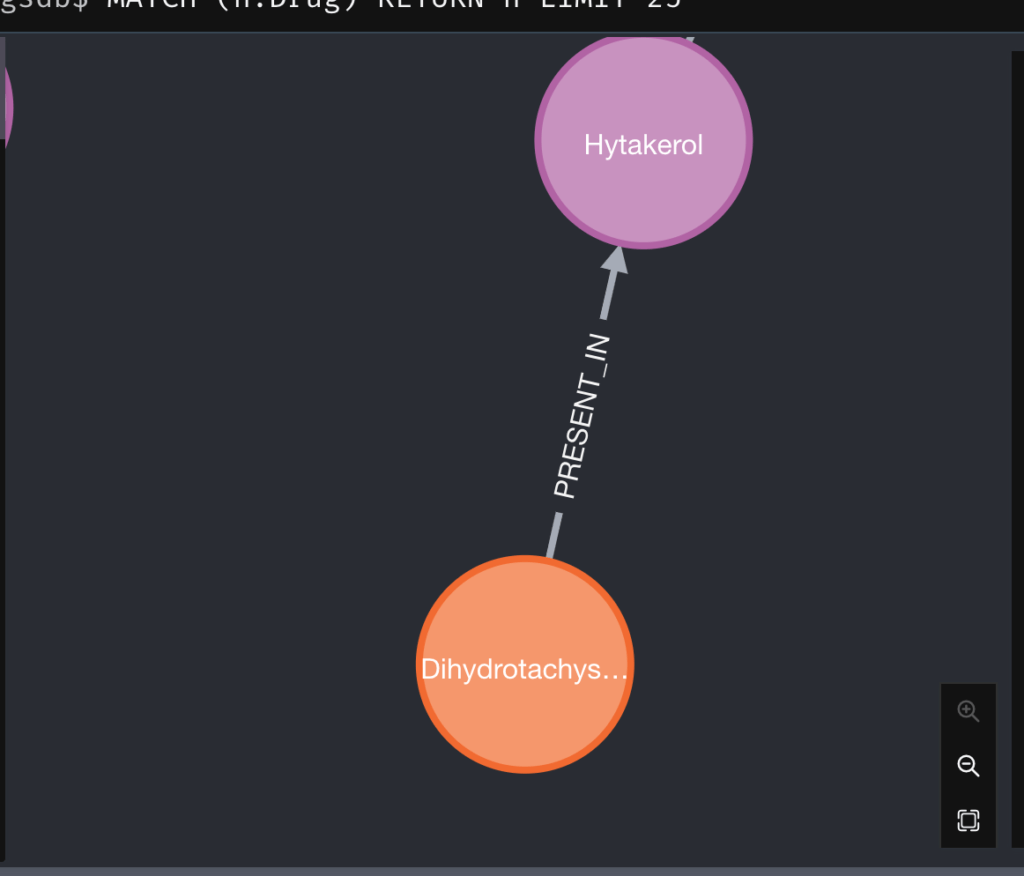
A relationship between an Ingredient and a Drug node looks like the below:
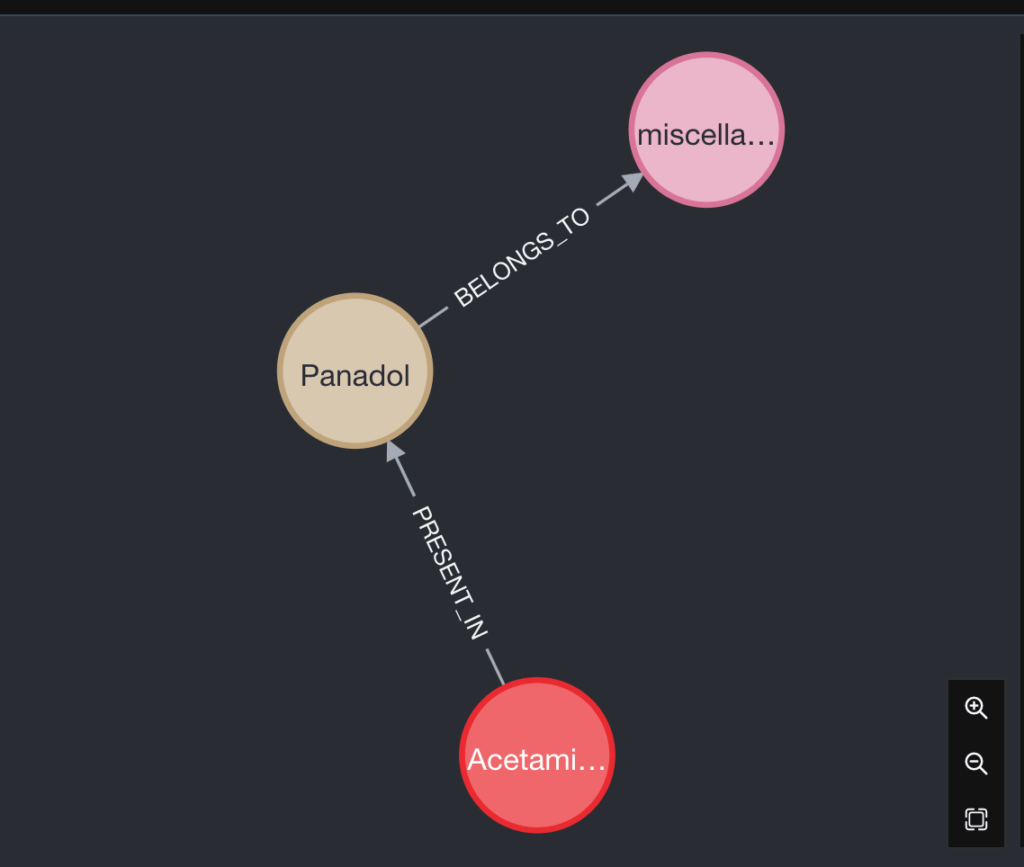
What If I want to know which DrugClass this Drug belongs to?
In simple words, Panadol consists of an Ingredient called Acetaminophen and it belongs to the drug class that deals with miscellaneous analgesics.
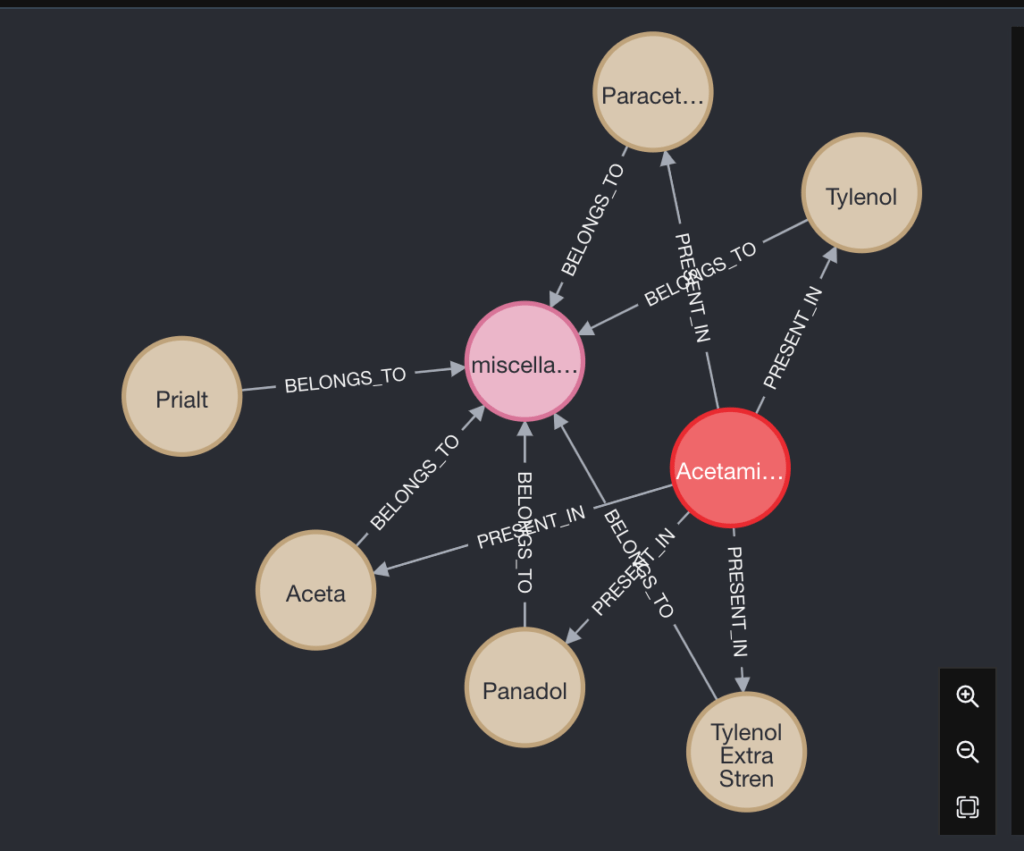
What if I want to see what other medicines act as a pain killer and belong to miscellaneous analgesics?
You may see some famous names like Tylenol and Paracetamol here. Cute, isn’t it? If I want, I can use this graph database to find alternative medicine if my desired medicine is not available.
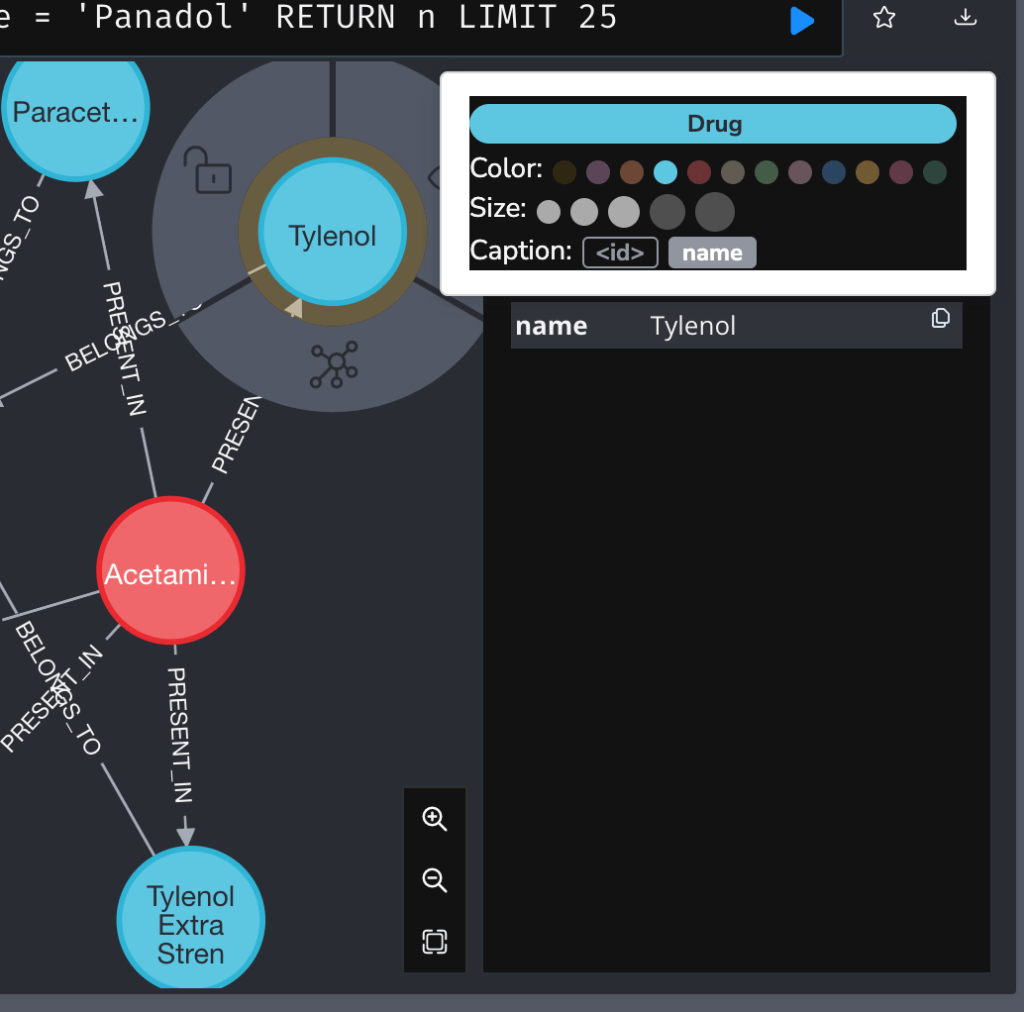
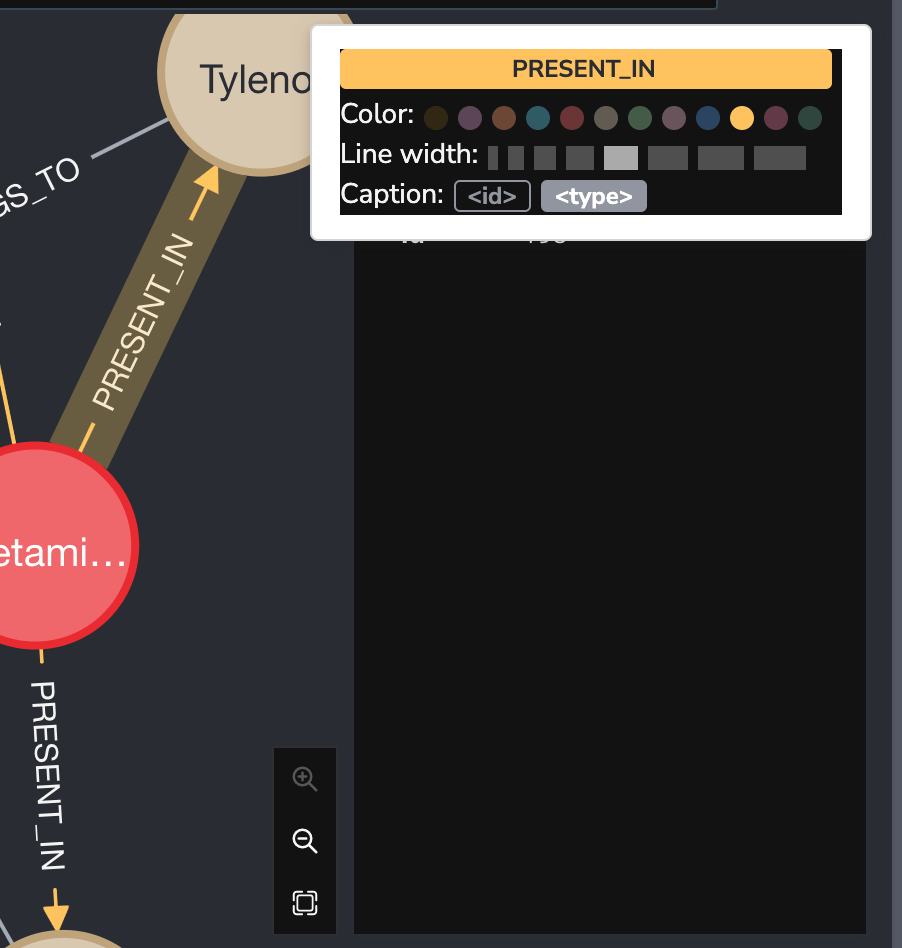
You can also change the color of a node.
So the color of relationship text.
You can reset to default by running :style reset common in the Neo4j browser. Also, if you want you can load the GraSS content from a url by running :style <URL>.
It is also possible to change the style programmatically. If you run the :style command in the browser pane, it shows the style that Neo4j terms as Graph Stylesheet (GraSS) in a window.
node {
diameter: 50px;
color: #A5ABB6;
border-color: #9AA1AC;
border-width: 2px;
text-color-internal: #FFFFFF;
font-size: 10px;
}
relationship {
color: #A5ABB6;
shaft-width: 1px;
font-size: 8px;
padding: 3px;
text-color-external: #000000;
text-color-internal: #FFFFFF;
caption: "<type>";
}
Pretty straightforward if you already know CSS.
As you notice, the label is not entirely visible. How about just increasing the node diameter? I explored thestyle.grass file, changed the diameter parameter to 60px and drag and drop to the same import pane. I also changed the arrow width.
The final changed .grass file looks like the below:
node {
diameter: 60px;
color: #A5ABB6;
border-color: #9AA1AC;
border-width: 2px;
text-color-internal: #FFFFFF;
font-size: 8px;
}
relationship {
color: #A5ABB6;
shaft-width: 2px;
font-size: 8px;
padding: 3px;
text-color-external: #000000;
text-color-internal: #FFFFFF;
caption: "<type>";
}
Conclusion
In this post, we explored how Neo4j helps to identify medicines that use certain active agents or discover further medicines in a certain drug class. Graph databases like Neo4j are not all about pharma or the medical field, you can use them for any kind of data that has certain relationships available. Like always, the code is available on GitHub.
If you like such posts then you may support my content and could help to buy a cup of ☕ 🙂